简介
细胞注释旨在通过分析细胞的基因表达谱,将细胞准确分类并确定其生物学身份 (如细胞类型、状态或亚群),从而解读数据的生物学意义、揭示组织或样本的细胞组成和功能异质性,并为包括细胞邻域分析、CNV分析、细胞通讯分析、轨迹分析、细胞空间距离关系分析、差异基因分析、转录因子分析在内的多种下游分析提供分类基础。
算法测评经验
- 如果希望快速获得结果,建议首先尝试使用Tangram或RCTD,对于没有单细胞参考的人类样本也可以先尝试SCimilarity。如果需要更准确的注释结果且有GPU资源,建议使用cell2location。详细算法测评信息,请参考此链接。
| 算法 | 性能 | 内存 | 时间 | GPU 加速 |
|---|---|---|---|---|
| cell2location | ⭐⭐⭐⭐⭐ | ⭐⭐ | ⭐ | 无 GPU 时极其慢 |
| RCTD | ⭐⭐⭐⭐ (排除未注释细胞) |
⭐⭐⭐ | ⭐⭐⭐ | |
| Tangram | ⭐⭐⭐ | ⭐⭐⭐⭐ | ⭐⭐⭐⭐ | CPU上速度较快,且可用 GPU加速 |
| SPOTlight | ⭐⭐ | ⭐ | ⭐⭐ | |
| SCimilarity | ⭐ | ⭐⭐⭐⭐⭐ | ⭐⭐⭐⭐⭐ | CPU上速度很快。可用 GPU加速,但效果不明显 |
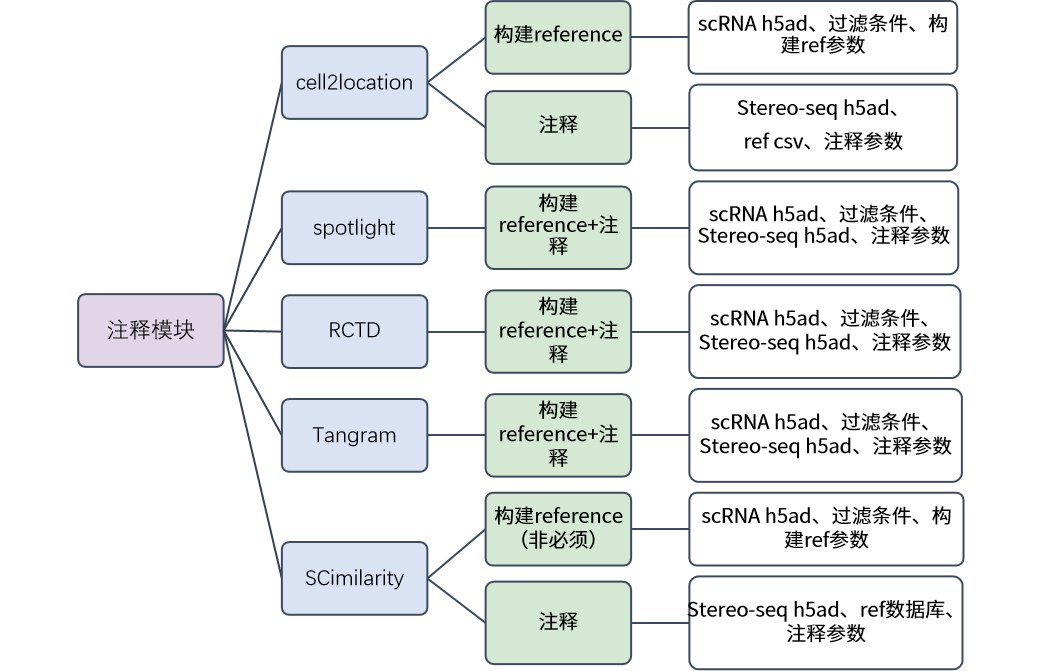
模块算法整体介绍
参考文献
- Biancalani, T., Scalia, G., Buffoni, L., Avasthi, R., Lu, Z., Sanger, A., ... & Regev, A. (2021). Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nature methods, 18(11), 1352-1362.
- Cable, D. M., Murray, E., Zou, L. S., Goeva, A., Macosko, E. Z., Chen, F., & Irizarry, R. A. (2022). Robust decomposition of cell type mixtures in spatial transcriptomics. Nature biotechnology, 40(4), 517-526.
- Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I., & Heyn, H. (2021). SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic acids research, 49(9), e50-e50.
- Heimberg, G., Kuo, T., DePianto, D. J., Salem, O., Heigl, T., Diamant, N., ... & Regev, A. (2025). A cell atlas foundation model for scalable search of similar human cells. Nature, 638(8052), 1085-1094.
- Kleshchevnikov, V., Shmatko, A., Dann, E., Aivazidis, A., King, H. W., Li, T., ... & Bayraktar, O. A. (2022). Cell2location maps fine-grained cell types in spatial transcriptomics. Nature biotechnology, 40(5), 661-671.